Scraping HTML with XPath
Scraping HTML
Internet pages provide a lot of information and often you would like to be able to access and manipulate it in another form than HTML: HTML is just plain verbose. What you would like is to get access to only the information you are interested in and get the results in a form that you can easily build more software. This is the objective of HTML scraping. In Pharo you can scrape web pages using different libraries such as XMLParser and SOUP. In this chapter we will show you how we can do that using XMLParser to locate and collect the data we need and JSON to format and output the information.
This chapter has been originally written by Peter Kenny and we thank him for sharing with the community this little tutorial.
Getting started
You can use the Catalog browser to load XMLParserHTML and NeoJSON just execute the following expressions:
Gofer it
smalltalkhubUser: 'PharoExtras' project: 'XMLParserHTML';
configurationOf: 'XMLParserHTML';
loadStable.Gofer it
smalltalkhubUser: 'PharoExtras' project: 'XPath';
configurationOf: 'XPath';
loadStable.Gofer it
smalltalkhubUser: 'SvenVanCaekenberghe' project: 'Neo';
configurationOf: 'NeoJSON';
loadStable.Define the Problem
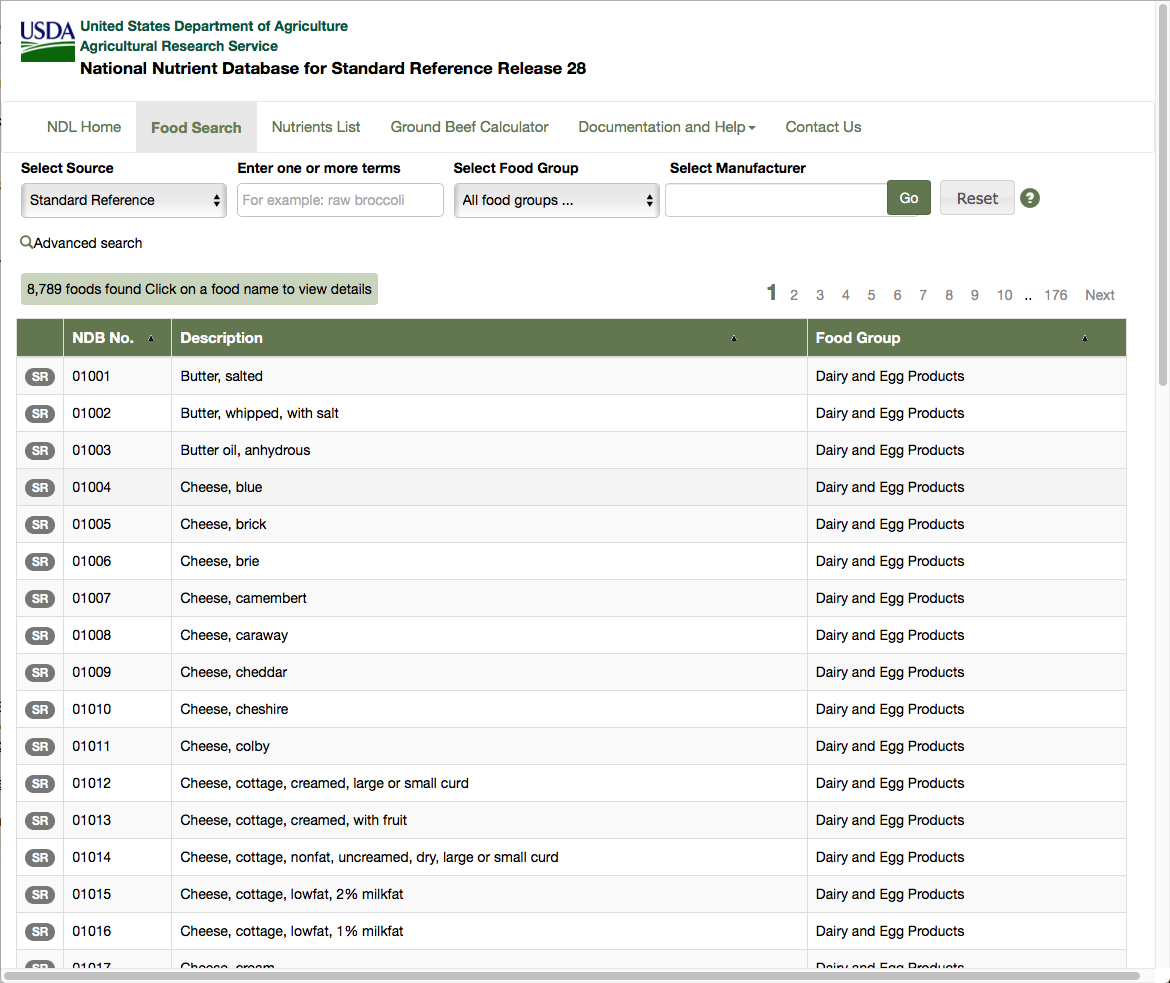
This tutorial is based on a real life problem. We need to consult a database published by the US Department of Agriculture, extract data for over 8000 food ingredients and their nutrient contents and output the results as a JSON file. The main list of ingredients can be found at the following url: https://ndb.nal.usda.gov/ndb/search/list?sort=ndb&ds=Standard+Reference (as shown in Figure ). You can also find the HTML version of the file in the github repository of this book https://github.com/SquareBracketAssociates/Booklet-Scraping/resources.
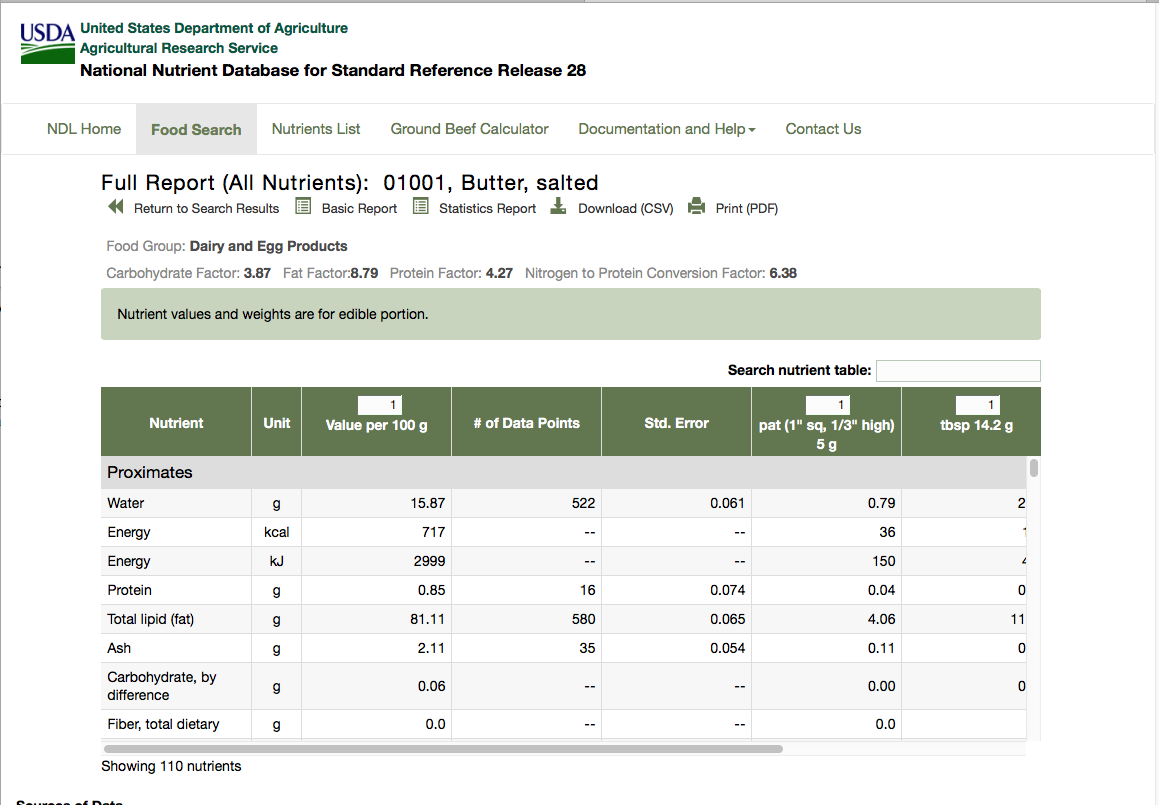
This table shows the first 50 rows, each corresponding to an ingredient. The table shows the NDB number, description and food group for each ingredient. Clicking on the number or description leads to a detailed table for the ingredient. This table comes in two forms, basic details and full details, and the information we want is in the full details. The full detailed table for the first ingredient can be found at the url: https://ndb.nal.usda.gov/ndb/foods/show/1?format=Full (as shown in Figure ).
There are two areas of information that need to be extracted from this detailed table:
- There is a row of special factors, in this case beginning with 'Carbohydrate Factor: 3.87'. This is to be extracted as a set of (name, value) pairs. The number of factors can vary; some ingredients do not have any.
- There is a table of data for various nutrients, which are arranged in groups - proximates, vitamins, lipids etc. The number of columns in the table varies from one ingredient to another, but in every case the first three columns are nutrient name, unit of measurement and quantity; we have to extract these columns for every listed nutrient.
The requirement is to extract all this information for each ingredient, and then output it as a JSON file:
- NBD number, description and food group from the main list;
- Factor names and values from the detailed table;
- Nutrient details from the detailed table.
First find the required data
To start, we have to find where the required data are to be found in the HTML file. The general rule about this is that there are no rules. Web site designers are concerned only with the visual effect of the page, and they can use any of the many tricks of HTML to produce the desired effects. We use the XML HTML parser to convert text into an XML tree (a tree whose nodes are XML objects). We then explore this tree to find the elements we want, and for each one we have to find signposts showing a route through the tree to uniquely locate the element, using a combination of XPath and Smalltalk programming as required. We may use the names or attributes of the HTML tags, each of which becomes an instance of XMLElement in the tree, or we may match against the text content of a node.
First read in the table of ingredients (first 50 rows only) as in the url.
| ingredientsXML |
ingredientsXML := XMLHTMLParser parseURL: 'https://ndb.nal.usda.gov/ndb/search/list?sort=ndb&ds=Standard+Reference'.
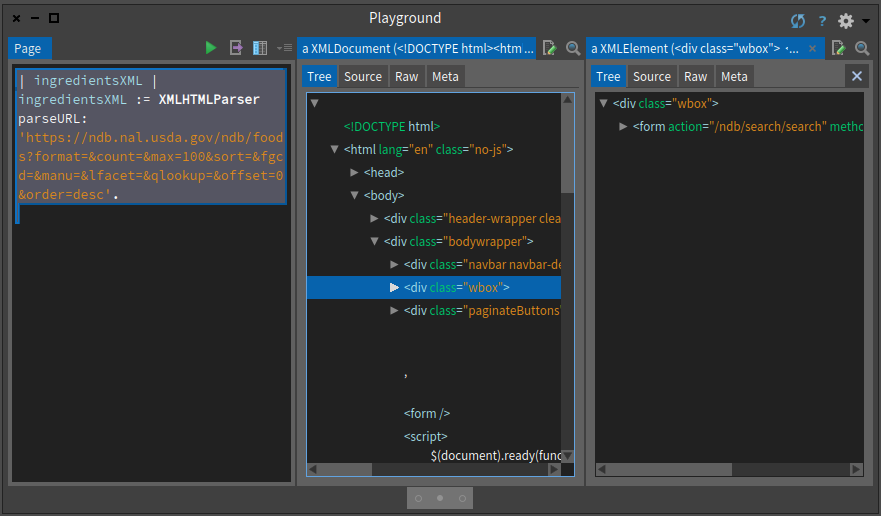
ingredientsXML inspectYou can execute the expression and inspect its result. You will obtain an inspector on the tree and you can navigate this tree as shown in Figure .
Since you may want to work on files that you saved on your disc you can also parse a file and get an XML tree as follows:
| ingredientsXML |
ingredientsXML := (XMLHTMLParser onFileNamed: 'FoodsList.html') parseDocument.
The simplest way to explore the tree is starting from the top, i.e. by opening up the <body> node, but this can be tedious and confusing; we often find that there are many levels of nested <div> nodes before finding what we want. Alternatively, we can use XPath speculatively to look for interesting nodes. In the case of the foods list, we might guess that the list of ingredients will be stored in a <table> node. Having parsed the web page as shown above in a playground, we can then enter:
ingredientsXML xPath: '//table'
and select 'do it and go', which shows an XMLNodeList of all the table nodes - only one in this case. If there were several, we could use the attributes of the node or any of its ancestors to locate the one we want. We find by searching up several generations a node <div class="wbox"> which is unique, so we could use this as a signpost. The table body contains a row for each ingredient; the first cell in the row is a "badge" which is of no interest, but the remaining three cells in the row are the number, description and group name that we want. The second and third cells both contain an emebedded node <a href="...."> showing the relative url of the associated detail table.
The exploration of the detail table proceeds in a similar way; we search for text nodes which contain the word "Factor", and then for a table containing the nutrient details. More of this below.
Going back to our problem
Here we present the essential points of the data scraping and JSON output for one item, in a logical order. The code is presented as it could be entered in a playground. There are brief comments on the format of the data and the signposts used to locate it. First read in the table of ingredients (first 50 rows only) as before.
ingredientsXML := XMLHTMLParser parseURL: 'https://ndb.nal.usda.gov/ndb/search/list?sort=ndb&ds=Standard+Reference'.The detail rows are in the body of the table in the div node whose class is 'wbox'.
ingredientRows := (ingredientsXML xPath: '//div[@class=''wbox'']//tbody/tr').
Note that the signposts do not need to show every step of the way, provided the route is unique; we do not need to mention the <table> node, because there is only one <tbody>. Now extract the text content of the four cells in each row; 'strings first' is a convenient way of finding the text in a node while ignoring any descendent nodes, and we routinely trim redundant spaces.
ingredientCells := ingredientRows collect:
[:row| (row xPath: 'td') collect:
[ :cell| cell strings first trim]].To prepare for export to JSON, it is handy to put the three required fields (ignoring the first) in a Dictionary indexed by their field names. Using an OrderedDictionary is not strictly necessary, but it does mean that the JSON output is easier for a human to understand.
ingredientsJSON := ingredientCells collect:
[ :row| { 'nbd_no' -> (row at: 2).
'full-name' -> (row at: 3).
'food-group' -> (row at: 4)}
asOrderedDictionary ].If we 'do it and go' the next line, we can see the JSON layout. For this demo, we do not need to export to a JSON file; it is easier to look at it as text in the playground.
NeoJSONWriter toStringPretty: ingredientsJSON first.We can find the relative url address of the ingredient details from the href in the second cell. Because this is the address of the basic details table, we edit it to discard all the parameters, so that we can edit in the parameters for the full table.
ingredientAddress := ingredientRows collect:
[ :row| (row xPath:'td[2]/a/@href') first value copyUpTo: $?].Up to this point, we have been constructing lists with data for all 50 ingredients in the table. To show how to process the ingredient details, we just process the first ingredient in the file. The production version would have to run through all the rows in the ingredientAddress collection. We read and parse the detail file, after editing the url.
ingredientDetailsXML := XMLHTMLParser parseURL: 'https://ndb.nal.usda.gov', ingredientAddress first, '?format=Full'.
The data for the factors are contained in <span> nodes within <div class="row"> nodes. This does not identify them uniquely, so we extract all such nodes with XPath and then use ordinary Smalltalk to find the ones mentioning the word 'Factor'.
factorCells := (ingredientDetailsXML xPath: '//div[@class=''row'']//span')
collect: [:each| each strings first trim].
factors := OrderedCollection new.
1 to: factorCells size by: 2 do: [ :index|
((factorCells at: index) matches: 'Factor') ifTrue: [factors addLast:
{'factor' -> (factorCells at: index).
'amt' -> ((factorCells at: index + 1) trimRight:[:c|c asInteger = 160])}
asOrderedDictionary]].Note: it appears that the web designers have used no-break space characters to control the formatting, and these are not removed by 'trim', so we use the 'trimRight:' clause above to remove them.
The layout of the nutrients table is messy, presumably to achieve the effect of the inserted row with the nutrient group name. This means that we cannot locate the nutrient rows using <tr> nodes, as we did for the main list. Instead we have to get at all the individual table cells in <td> nodes, and then count them in groups equal to the row length. Since the row length is not a constant, we have to determine it by examining the data for one row that in a <tr> node.
nutrientCells := (ingredientDetailsXML xPath: '//table//td') collect: [:each|each strings first trim].
nutRowLength := (ingredientDetailsXML xPath: '//table/tbody/tr') first elements size.
nutrients := OrderedCollection new.
1 to: nutrientCells size by: nutRowLength do:
[:index|nutrients addLast:
{ 'group' -> (nutrientCells at: index).
'nutrient' -> (nutrientCells at: index + 1).
'unit' -> (nutrientCells at: index + 2).
'per100g' -> (nutrientCells at: index + 3) }
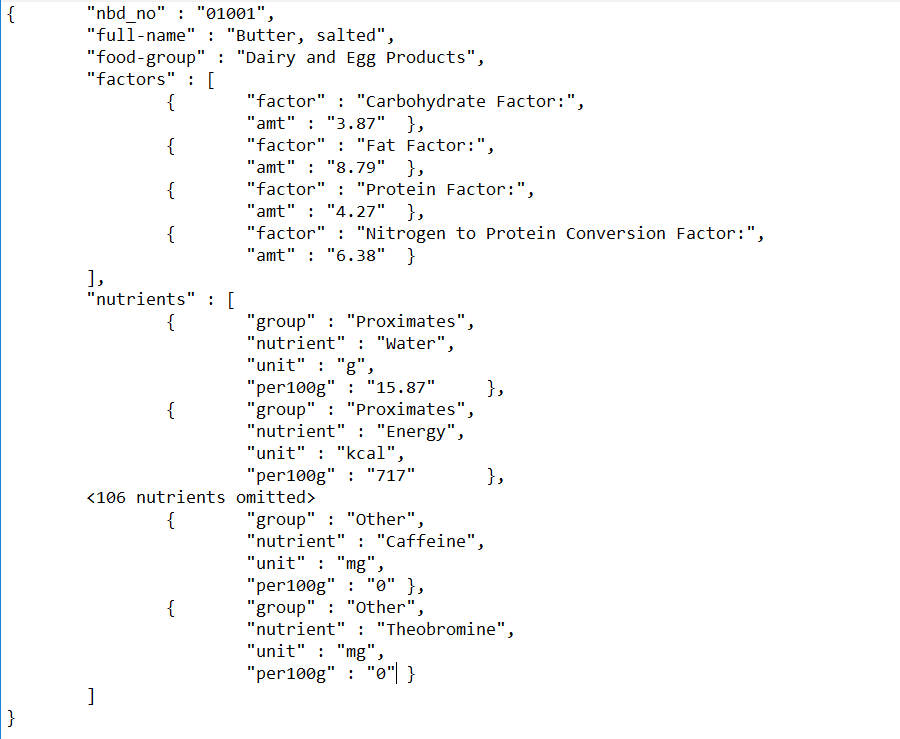
asOrderedDictionary ].Finally assemble all the information for the first ingredient as a JSON file. NeoJSON automatically takes care of embedding dictionaries within a collection within a dictionary. (See specimen in Figure )
NeoJSONWriter toStringPretty:
((ingredientsJSON first)
at: 'factors' put: factors asArray;
at: 'nutrients' put: nutrients asArray;
yourself).Turning the pages
The code above will extract the data for one ingredient, and could obviously be repeated for all the 50 items in one page of data. However, the entire database contains 8789 ingredients at the time of writing, which amounts to 176 pages. The database seems to impose a limit of 50 ingredients per page, so to process the entire database we need to read the pages in succession. Each page contains a link which, if clicked, will load the next page. We can do this programmatically, by finding the link after processing the page. The link is contained in node <div class="paginateButtons">, so we can use the code:
nextButtons := (ingredientsXML xPath: '//div[@class=''paginateButtons'']//a')
select:[:node| node strings first = 'Next'].
nextURL := (nextButtons size > 0)
ifTrue:['https://ndb.nal.usda.gov', (nextButtons first attributeAt: 'href')]
ifFalse: [nil].This is a common requirement in processing large databases on the web, and so we can use a standard pattern:
<code to initialise results>
nextURL := <url for first page of database>
[nextURL isNil] whileFalse:
[pageXML := XMLHTMLParser parseURL: nextURL.
<code to extract data from pageXML to results>
<code to determine nextURL from pageXML; should yield 'nil' for last page>
]Conclusion
We have presented a way to extract information from a structured document. The methods used are of course particular to the layout of the USDA database, but the general principles should be clear. A mixture of XPath and Smalltalk can be used in order to locate the required data.
One problem which can arise, if we need to repeat the extraction with updated data, is that the web designers can change the layout of the pages; this did in fact happen with the USDA table in the 15 months between originally tackling the problem and writing this article. The usual result is that the signposts no longer work, and the XPath results are empty. If the update is being run automatically, say on a daily basis, it may be worth while inserting defensive code in the processing, which will raise an exception if the results are not as expected. How to do this will depend on the particular application.